Written by Emil O. W. Kirkegaard.
There are two exciting pieces of news in embryo selection. The technology is finally moving forward at a steady clip. Maybe we aren’t doomed by dysgenics after all!
First off, Herasight — the new startup for embryo selection I covered in July — just published their follow-up validation study concerning the prediction of intelligence: ‘Within-family validation of a new polygenic predictor of general cognitive ability’.
The authors present a new polygenic score that, quote, “demonstrates a substantial increase in predictive accuracy both among unrelated individuals and within-families relative to existing predictors”. Specifically, their polygenic score explains 16% of the variance in general cognitive ability in the UK Biobank, corresponding to a standardized effect of β = 0.41. What’s more, when they tested it in a sample of sibling pairs, they obtained an estimate that was only slightly lower — indicating minimal attenuation within families. The authors replicated these results in the Adolescent Brain and Cognitive Development dataset. After correcting for reliability, their polygenic score correlated with latent cognitive ability at r = .45, an impressive result. It even achieved good performance in non-European samples, retaining 64% of its magnitude in a sample of black Americans.
The main focus of the paper is investigating two issues that complicate polygenic score validation: within-family comparisons (i.e., sibling comparisons) and the role of measurement error. Table 1 gives the key results:
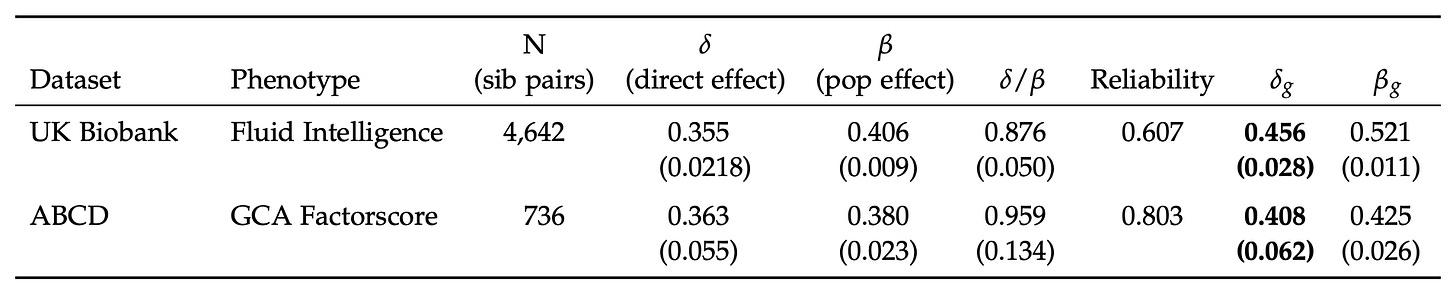
Using their proprietary model (based on SBayesRC) they achieved out-of-sample between-family effects of about 0.40 and within-family effects of about 0.36. The ratio between these numbers is important here since it tells us how much of the validity remains when controlling for factors that are shared by siblings.
Since embryo selection involves choosing among siblings, between-family validity is irrelevant. When a polygenic score works much less well within families than between them, it suggests there is non-genetic confounding in the models, or something more complex like gene–gene or gene–environment interactions or perhaps parental genes that boost the intelligence of children but only if they also have those genes. The ratio Herasight achieved is about 0.90, meaning that their polygenic score worked almost 90% as well within families as between them. Actually, given that the effect of measurement error is enhanced in sibling studies (because the model is relying on difference scores, each of which has its own error component), the true ratio could be close to 1.
These are great results. But the authors go further and consider the classical test theory approach to polygenic scores, something I’ve been advocating for years. Given that the measures of intelligence in these samples are not perfect (they are never perfect), the standardized effects will, again, be smaller than their true values. Thus, if you can estimate the size of this reduction from the reliability (the correlation of a test with itself taken again), you can estimate the true standardized effect. The authors did this in both of their samples.
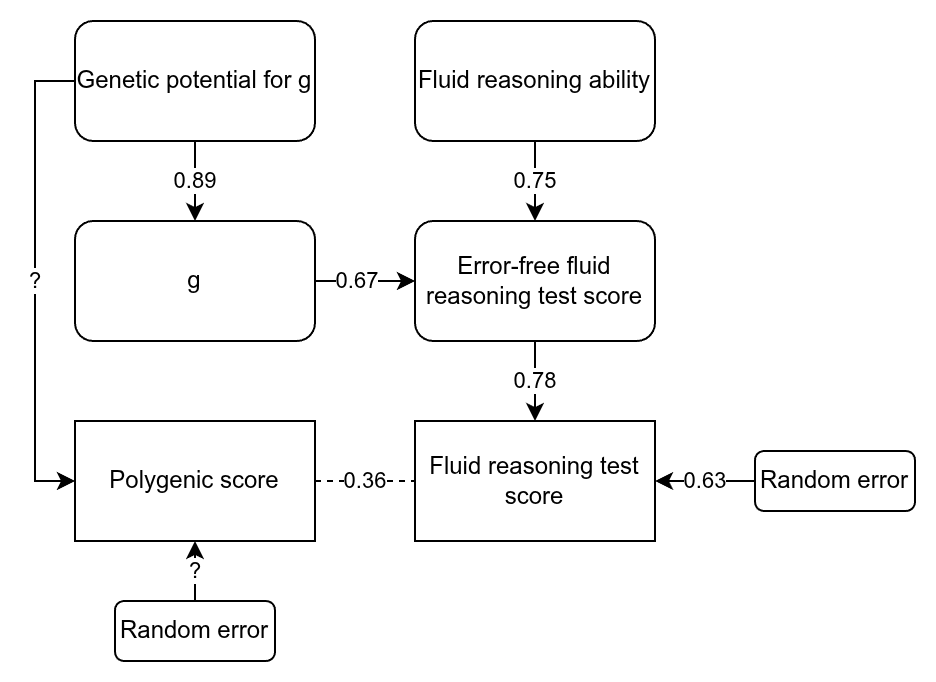
Here I want to nitpick regarding the UK Biobank data. Herasight used just the fluid intelligence test (because it has the largest n), which is in fact quite poor, being based on only 13 items. You can find the items here if you are curious. They include questions like “Add the following numbers together: 1 2 3 4 5 – [What] is the answer?” and “If Truda’s mother’s brother is Tim’s sister’s father, what relation is Truda to Tim?”. A subset of participants took this particular test twice, and their scores correlated at 0.607 (the reliability seen in the table above). Dividing the standardised effects by the square root of the reliability produces the estimated true standardised effects, given in the last column of the table. So what’s my complaint? The fluid reasoning test in the UK Biobank is rather limited as an intelligence test, and cannot be assumed to be perfectly g-loaded. Here’s a drawing to illustrate:
We can think of the raw score on the test as being caused by three variables we don’t observe directly: 1) general intelligence, 2) non-g ability related to fluid reasoning, and 3) random error. A study of the UK Biobank tests showed that the fluid test has a g-loading of 0.52. This is without taking into account measurement error. Using the quick method of adjusting this value for reliability (i.e., assuming that the factor structure doesn’t change) gives an error-free g-loading of 0.67. As such, the non-error causes of performance on this test must have a loading of 0.74. In other words, only about half the error-free variation on this test is due to g, with the rest being due to other abilities, including familiarity with IQ testing. In the drawing above, I’ve also filled out some other paths, assuming a heritability for g of about 80% (their twin estimate for ABCD was 64%, which when corrected becomes 80%).
What all this means is that when they find that they can predict the error-free variation in the fluid intelligence test with a standardized effect of 0.46 or so, this is a mixture of g and non-g factors. The problem does not arise in the ABCD dataset because it includes a battery of 10 tests of varying nature, meaning that the general factor is very close to being just g plus error. This may explain some minor oddities in the authors’ results, such as that the error-free population effect sizes do not overlap in their confidence intervals. Note that non-g abilities can also be quite heritable, particularly when error is removed statistically, so just because you predict something heritable doesn’t mean it is only g.
Next, the authors consider the effect of error on heritability estimates using the “all-SNPs” approach. Since they’re dealing with variance, the correction is to divide by the reliability itself, not the square root. They do some simulations to show that this method works correctly. Strangely, they don’t seem to actually apply this to their SNP-based estimates. However, they do report that the SNP-based estimate for the fluid intelligence test is 23%, which becomes 38% after correction. They apparently did not report the SNP-based estimate for the ABCD dataset. There are many studies analyzing the ABCD, and it’s possible that one of them has reported it.
Anyway, Herasight’s SNP-based heritability is an estimate for an additive, linear polygenic score trained on the relevant SNPs but with infinite sample size. The polygenic score’s actual heritability is about 0.43^2 = 18%. As such, we could say that the current PGS captures about half the variance possible given the constraints (i.e., these particular genetic variants). However, the polygenic score’s practical utility for selection is a function of the standardized effect, not the r² or h² values.
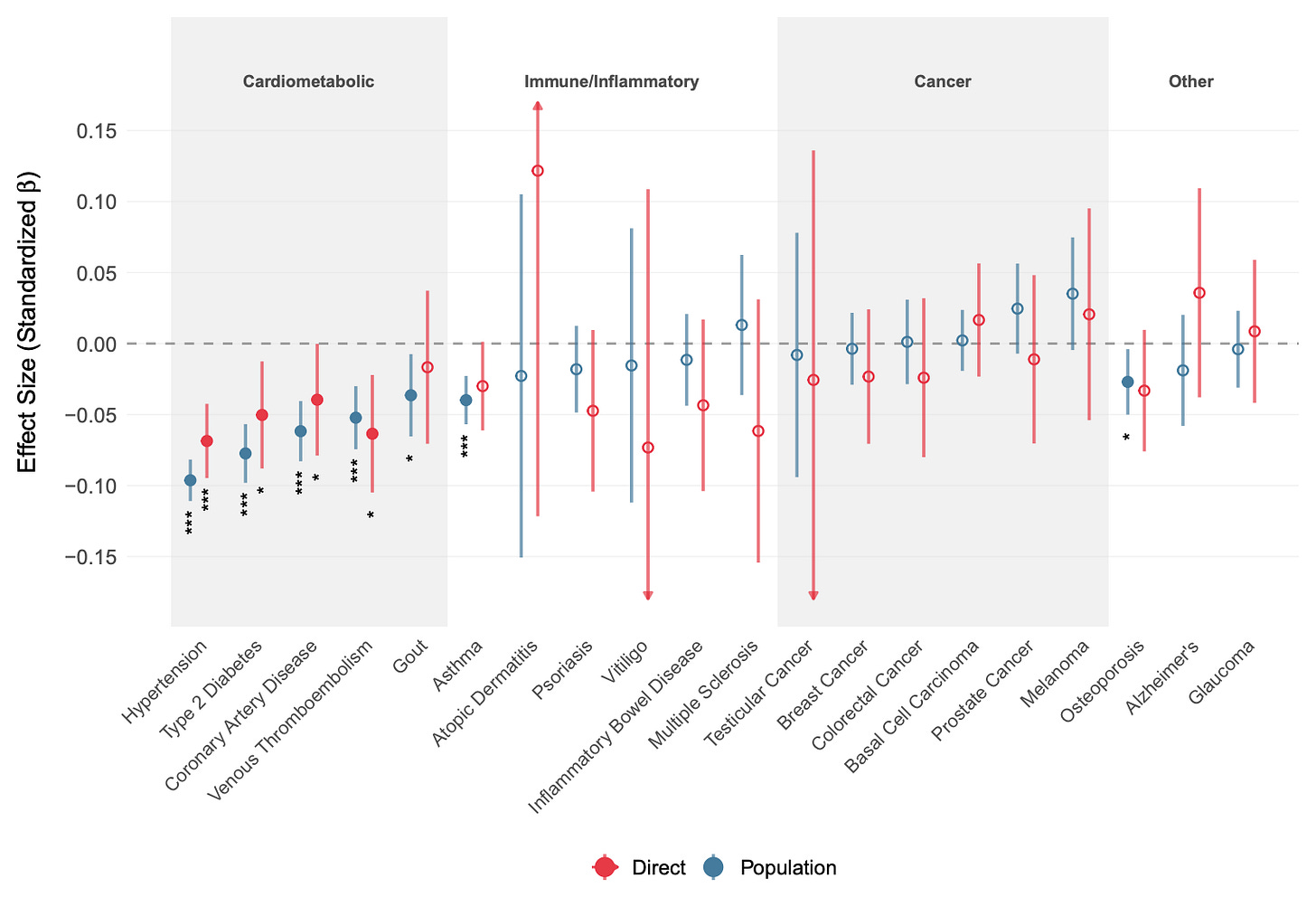
Moving on to the strangest part about the study: the polygenic score’s association with disease outcomes.
These are standardized effects for other outcomes, mainly diseases. It is known that intelligence is mildly protective against a variety of diseases and that this effect is mainly genetic in origin. As such, selecting for higher intelligence essentially gives you “freebies”. The strange thing is that Alzheimer’s disease shows just about no effect. Why strange? Well, large national studies show that intelligence is protective against this particular disease, and therefore any polygenic score for intelligence should be too.
The authors also looked at their polygenic score’s relationship to other psychological measures in the ABCD dataset:
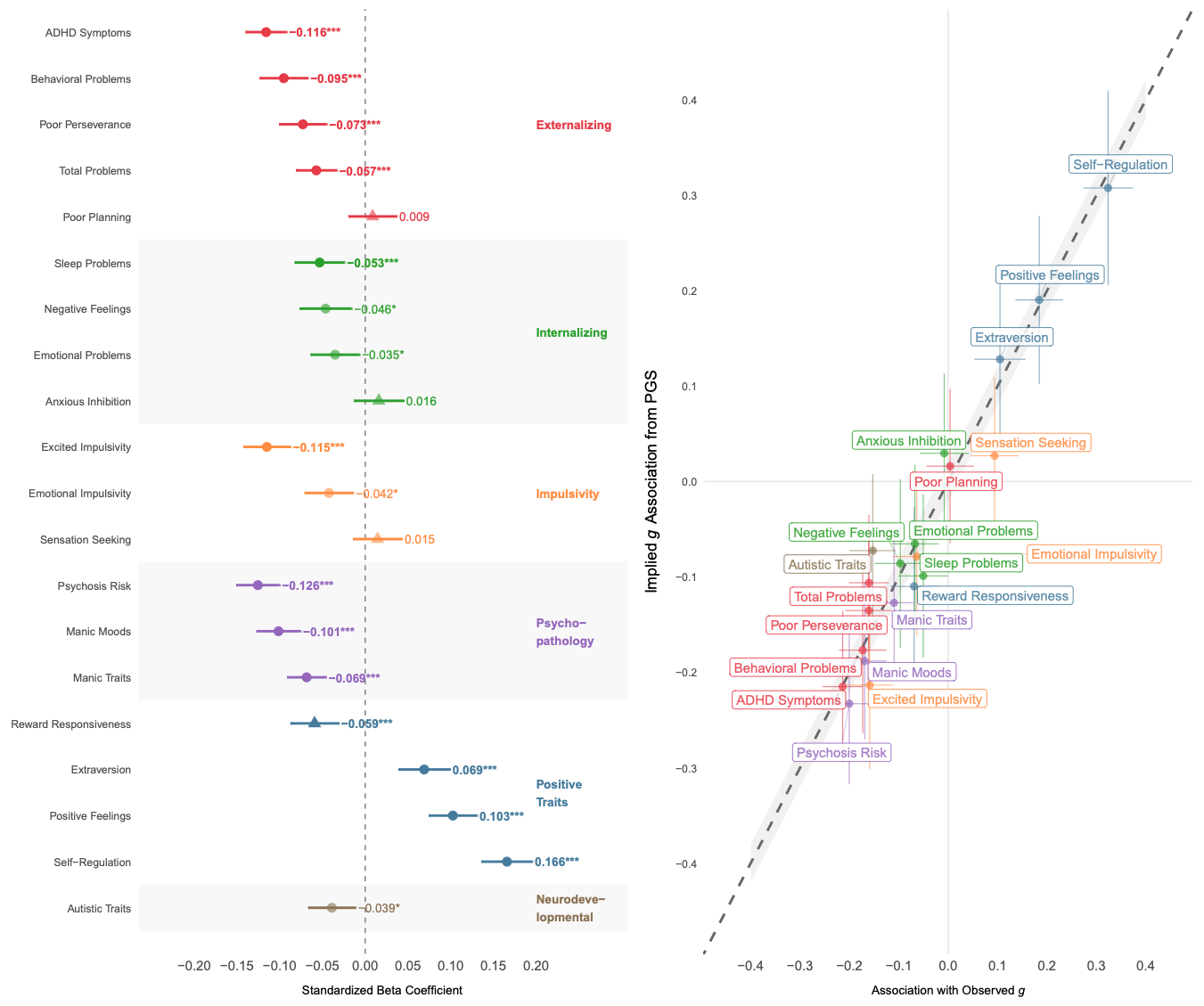
Since the genes for many traits overlap, selecting for one trait also means selecting, in part, for and against certain other traits. Whether this is good or bad depends on the directions of these correlations and how we value the traits. In this case, most people prefer for their children not to have ADHD, and, fortunately, the correlation is negative. Most of the other correlations for mental health conditions were negative too. Plus, there were positive correlations for traits that are generally valued, including extroversion. This means that selecting for intelligence using this polygenic score would also give you slightly more extroverted children.
Herasight’s polygenic score also had a slight negative correlation with autism. This is curious because many studies find small positive genetic correlations between polygenic scores for intelligence/education and autism. This may be because we’re dealing with a children’s sample, or because autism has become “more popular” in recent generations, making it less elite. Who knows.
Meanwhile, the other new piece of embryo selection news concerns the startup Nucleus Genomics. It appeared that they’d decided to publish their disease predictor models, which would have been an unexpected and welcome move towards open source genomics. But it turns out that it’s actually one of those fake “open access” arrangements where you have to submit an application. I gave it a try! It was at least less onerous than the usual academic ones — so maybe still an improvement.
Nucleus Genomics have trained their models on the UK Biobank and All of Us datasets, giving a total sample size of maybe 1.5M. This means that Herasight and other parties can now (in theory) download the models and test them against their own models. Better yet, someone should set up an independent institution for validating genetic models.
It should collect a new sample with measures of the relevant traits. And it should run in-house validations of published genetic models. This way, no one can cheat and overfit their models: the institute could provide everybody with unbiased validation results, just as we have third parties ranking existing AIs on a private IQ test the AIs have never seen. Many countries are building or expanding their biobanks, so it wouldn’t be particularly difficult for them to set aside a few thousand participants for such an institute’s internal dataset. If a few countries did this, the internal validation dataset could quickly reach 10K, including siblings for the crucial within-family validation studies. What’s not to like about this idea?
Maybe someone reading this article can try to move forward with this idea. The USA, for instance, is collecting a lot of data for their Million Veteran Program. This dataset is not used much by academics due to security clearance issues relating to the paranoia of the military. However, Team Trump could in theory just tell the military to cut it out, and to provide us with data for a genetic prediction model “clearing house” in the public interest.
Emil O. W. Kirkegaard is a social geneticist. You can follow his work on Twitter/X and Substack.
Consider supporting Aporia with a paid subscription:
You can also follow us on Twitter.




"There are two exciting pieces of news in embryo selection. The technology is finally moving forward at a steady clip. Maybe we aren’t doomed by dysgenics after all!"
That is good news. Long-term, I am hopeful that genetic enhancement will advance humanity.
"This may be because we’re dealing with a children’s sample, or because autism has become “more popular” in recent generations, making it less elite. Who knows."
I believe that it is most likely that the spectrum for autism has been broadened over the years.
Thanks for the interesting article.
Isn’t the 9 point advantage from 10 embryos roughly what Gwern estimated was possible with much older data? What’s the game changer in this story?